ADPenetrance: penetrance estimation for autosomal dominant traits
Penetrance is the probability of developing a particular trait given that a person harbours a certain genetic variant or set of variants. This tool uses population-scale data to allow estimation of the penetrance of risk variants associated with autosomal dominant traits. These estimates are derived from:
- the rate at which one of several 'disease states' occurs among people who harbour the variant.
- the size of families (sibships) within the studied population.
The tool is operated within the 'Penetrance Calculator' tab
This approach to penetrance calculation was initially described within Spargo et al. (2021). The disease model upon which it is based is presented within Al-Chalabi and Lewis (2011). The method is also available as an R function, hosted on GitHub.
On this tab, we outline: (1) the assumptions of this method, (2) the main operations of the tool, (3) information regarding the integrated data repository for approximating sibship size.
Assumptions
Disease model assumptions
- All individuals harbouring the variant are ascertained.
- All variants are inherited from exactly one parent; homozygosity is not included within the model and there are no de novo variants.
- A nuclear family containing a parent who harbours the variant can be classed into one of three disease states:
- familial - more than one family member is affected.
- sporadic - one family member is affected.
- unaffected - no family members are affected.
- Penetrance is not complete.
Additional assumptions for penetrance calculation
- Individual families are represented only once across the sample.
- Weighting factors, average sibship size, and disease risk for people not harbouring a variant (which defaults to 0) are absolute values.
- The value specified for sibship size is representative of sibship size across disease state groups.
- People can also be described within an ‘affected’ disease state, characterised by the instance of one or more family members being affected (i.e. this does not stratify the affected population between the familial and sporadic states).
- Disease state classifications are assigned assigned according to the status of the sampled person and first-degree relatives only
- When sampling across only families where the variant occurs, disease state classifications for sampled families will not change at a future time
- When estimating variant frequencies within disease states across cohorts of people with and without the variant, family disease states will change comparably over time for people with and without the variant
- Unless an estimate of disease risk for people not harbouring a variant is provided, those not harbouring the variant are unaffected.
Workflow
Step 1: Estimating disease state rate (the rate of state X)
To calculate penetrance via this method it is necessary to calculate the rate at which one of the disease states represented in the available data is observed among people who harbour the assessed variant across all of those disease states represented. In this tool, the user can select to estimate this via one of three methods. In formats (1) and (2), this calculation is performed within the tool based on data given by the user. In format (3) we allow the user to specify this estimate directly for a given state, as requested by the calculator. In formats (1) and (2) the user should indicate the frequency at which the assessed variant occurs in either two or three of the 'familial', 'sporadic' and 'unaffected' states or for the 'affected' and 'unaffected' states (see assumptions for state definitions). In format (1) these are given as variant counts and sample size, and in format (2) as variant frequencies. The user must then also specify the disease characteristics requested, including the rate at which familial/sporadic disease occur among the affected population and the overall population risk of being affected. Which disease characteristics are required depends upon the disease states for which variant frequency estimates have been given; the tool will indicate which should be specified.
If data are provided in formats (1) or (2) they will be used to calculate, as a weighted proportion, the rate at which one of the disease states occurs among the states for which variant frequency data are provided. This is the observed rate of 'state X', where state X denotes one of the states for which variant frequency data were given. If data are provided in format (3), the user should specify the rate of state X among people harbouring the assessed variant across those represented disease states, as requested by the calulator. If the familial state is represented within input data, then state X is familial. If only the sporadic and unaffected states are represented then state X is sporadic. If the affected and unaffected states are represented, then state X is affected. Note that the affected and unaffected states represent cases and controls respectively.
The result of this calculation represents the 'observed' rate of state X shown in the output Table.
Step 2: Generating a lookup table
The user must specify the average sibship size within the population from which the variant frequency estimates were drawn. We recommend also specifying the disease risk who do not harbour the variant, which is important for accurate penetrance estimation in common traits (e.g. where this risk is >0.01); by default this risk is assumed to be 0. The tool will produce a sequence of potential penetrance values from 0 to 1, increasing at increments of 0.0001.
Using the extended Al-Chalabi and Lewis (2011) disease model, according to the specified sibship size, and disease risk for people not inheriting the variant, and for each value of penetrance, the tool calculates the probabilities of families harbouring the variant presenting as unaffected, familial and sporadic. From these disease state probabilities, taking only those states for which data were provided in Step 1, the tool will calculate the 'expected' rate of state X at each penetrance value. A lookup table is then generated, storing each value of the expected rate of X (being either familial, sporadic, or affected) alongside the penetrance value to which it corresponds. When X is the affected state, the expected affected rate is determined as the sum of the expected familial and sporadic rates at a given penetrance value.
Step 3: Querying the lookup table
The lookup table is queried to identify the expected disease rate of X closest to the observed rate of X obtained in Step 1. The corresponding penetrance estimate within the lookup table is then obtained. Please note that the estimate here is subject to a systematic bias underlying the approach, and must be adjusted in Step 4 to obtain the final penetrance estimate.
Step 4: Adjust estimate for systematic bias
The penetrance estimate obtained in Step 3 is adjusted in this step, to account for systematic biases present. The degree of correction is determined by penetrance value estimated and the error in that estimate predicted under a polynomial regression model. This regression model is fitted based on errors in Step 3 penetrance estimates observed in a simulated dataset which is generated according to the states used to model the rate of state X, the mean sibship size and disease risk for people without the variant, as defined in the input data. The simulated dataset contains a representation of the distribution of sibship sizes across sampled families, following a Poisson distribution where lambda is the mean sibship size defined. Penetrance is then estimated for this population as in Steps 1-3 across a series of true penetrance values between 0 and 1, and the difference between true penetrance values and estimates is determined: estimate Error = true penetrance value - estimated penetrance. An nth degree polynomial regression model, between 1 and 5 degrees, is then fitted to these data, selecting the best-fit model according the Akaike Information Criterion. The penetrance estimate obtained for the real sample data is then adjusted according to error predicted in this estimate within the fitted model: adjusted penetrance estimate = unadjusted estimate + predicted error.
OPTIONAL: Error propagation
Confidence intervals can be calculated for the penetrance estimate based on error terms for the variant data provided by the user. Data format options are described in the 'Tool operation guide' provided in the Penetrance Calculator tab. If data for Step 1 are given in format (1), then error propagation is performed by default. If data are provided in formats (2) or (3) then the user can opt to provide error terms for these estimates, either as standard errors or as confidence intervals, and then indicate the desired confidence level for the resulting penetrance estimate. When input data are provided in format (1) or format (2) with error terms, the calculus-based approximation of error propagation (Hughes & Hase, 2010) is applied to obtain confidence intervals for the estimate of the observed disease state rate. When input data are provided in format (3), the standard error of the disease state rate is taken as specified, or converted into the standard error from confidence intervals using z-score conversion. Any transformations between confidence intervals and standard errors are performed using z-score conversion, for the confidence level indicated by the user. The lookup table constructed within Step 2 is queried as in Step 3 for the upper and lower bounds of the observed disease state rate and corresponding penetrance estimate, which is then adjusted as in Step 4 to determine confidence interval bounds the adjusted penetrance estimate.
Sibship data repository
Sibship size is a key parameter in the Al-Chalabi & Lewis (2011) model, determining disease state probabilities at each potential penetrance value.
If known, the user can manually define the average sibship size for the sample. We also provide a dataset of Total Fertility Rate estimates for many world regions, at a country level or aggregated across multiple countries, obtained from the latest data available within the World Bank database. Total Fertility Rate represents the number of children that would be born to a woman if she were to live to the end of her childbearing years and bear children in accordance with age-specific fertility rates of the specified year. It can be applied as a proxy for average sibship size in the population of interest.
The penetrance calculator tool is available here - please see below for information regarding tool operation and each data field
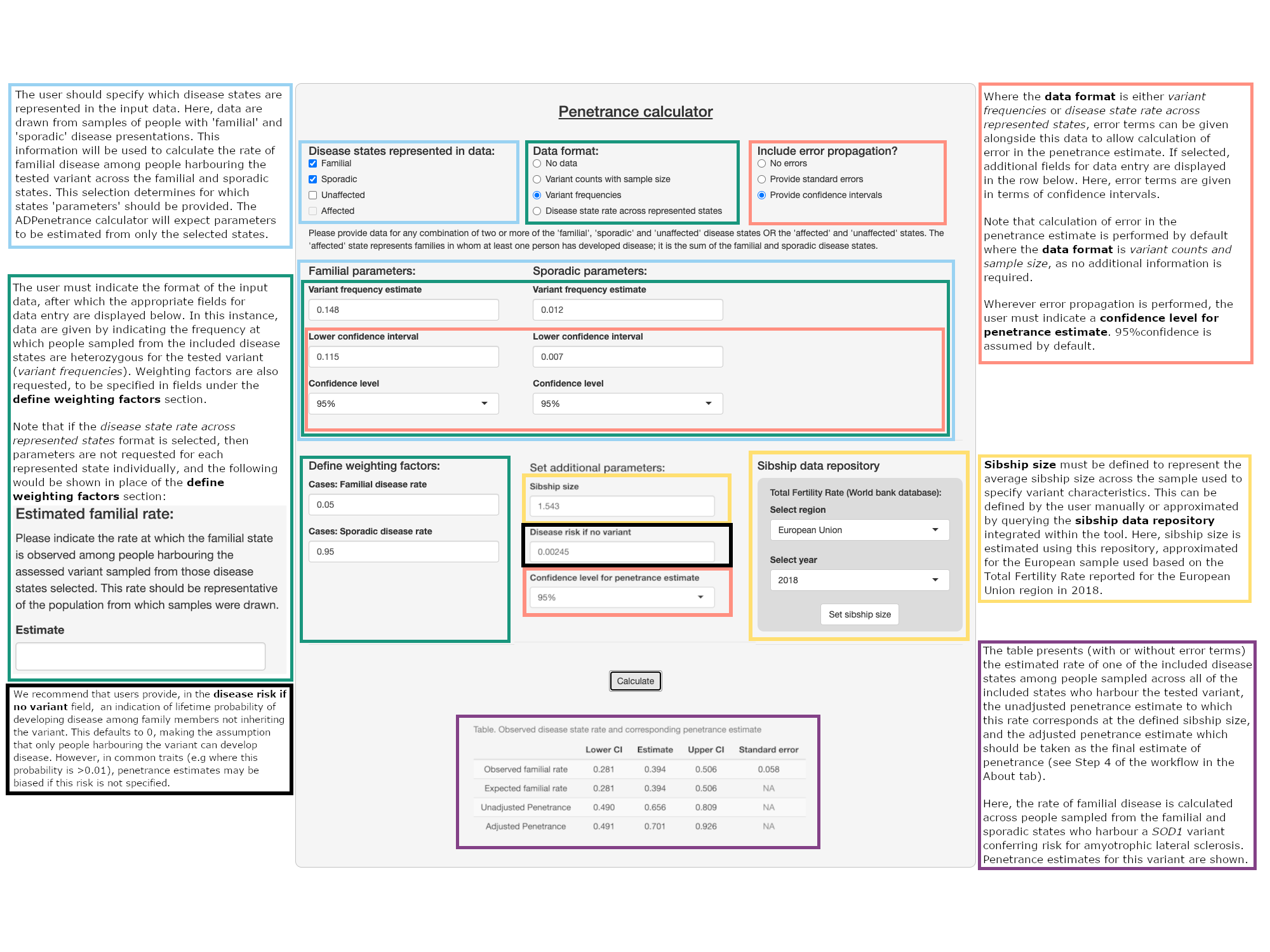
Penetrance calculator
Disease states represented in data:
Data format:
Include error propagation?
Please provide data for any combination of two or more of the 'familial', 'sporadic' and 'unaffected' disease states OR the 'affected' and 'unaffected' states. The 'affected' state represents families in whom at least one person has developed disease; it is the sum of the familial and sporadic disease states.
Familial parameters:
Sporadic parameters:
Affected population parameters:
Unaffected population parameters:
Define weighting factors:
Set additional parameters:
Sibship data repository
Total Fertility Rate (World bank database):
Tool operation guide
To operate this tool, the user must indicate in the disease states represented in data field for which disease states variant characteristics have been assessed. They should check either two or more of the 'familial', 'sporadic' and 'unaffected' disease states, or the 'affected' and 'unaffected' states. Note that the unaffected state represents control populations. The familial state represents cases in whom a first-degree relative is also affected by the trait, while the sporadic state represents cases without an affected first-degree relative. The affected state encompases the familial and sporadic states and does not stratify cases according to the number of affected first-degree relatives.
In the data format field, the user should indicate whether they will provide data on variant characteristics as (1) Variant counts with sample size, (2) Variant frequencies, or (3) Disease state rate across represented states:
- The user must indicate the number of people who harbour the assessed variant within samples of suitably sequenced people across each of the represented disease states
- Variant characteristics in each represented disease state, equivalent to those required in data format (1), should be provided as a proportion
- The user must provide an estimate of the rate at which the requested disease state (shown in the tool) is observed among people who harbour the assessed variant among those disease states sampled
Error propagation is performed by default if data are provided in format (1). If either format (2) or (3) are selected, the user should additionally indicate whether to include error propagation in the field displayed on the right. This is done by selecting whether error terms for the estimate(s) provided will be given as standard errors or as confidence intervals.
Once the above options are specified, the user can input values necessary for penetrance calculation into the numeric fields shown. Those displayed will vary according to the data format chosen and the disease states represented.
Displayed if data are provided in either formats (1) or (2)
Familial/sporadic/unaffected/affected parameters: The user must provide variant information in each of the represented disease states.
Where data format (1) is selected, only the variant counts and sample size details need to be specified.
Where data format (2) is selected, these should be provided as proportions. If the user has selected to perform error propagation, they should additionally specify either the standard errors or the lower confidence interval of the estimates provided, as previously indicated. If confidence intervals are given, the confidence level of this interval must be selected from a list of options; 95% confidence is assumed by default. Note that although the input can accept standard error or confidence intervals, the penetrance estimate output will always be a confidence interval (at the confidence level specified in the confidence level for penetrance estimate field).
Define weighting factors: The user should specify the weighting factors requested by the tool. These will be taken alongside the variant frequencies provided to derive the 'observed' familial/sporadic/affected disease state rate.
The cases: familial/sporadic disease rate factors indicate the rates of familial and sporadic disease among the affected population. The familial rate is the frequency at which people affected by the disease have an affected first-degree relative, while the sporadic rate is the frequency at which affected people do not have an affected first-degree relative (1 - familial rate). This information is used to weight variant frequency estimates given for the familial/sporadic disease states and ensures that the variant frequency estimates are scaled to the proportion of the affected population which each state represents. A value of 0.5 is given by default, representing an equal weighting. The population: disease risk factor indicates the likelihood of a general population member becoming affected by a particular age. This could be the lifetime incidence, for example. It is used to adjust variant frequency estimates in analysis featuring the unaffected disease state.
Displayed if data are provided in format (3)
Estimated familial/sporadic/affected rate: The user should indicate the rate at which the familial/sporadic/affected state is observed among people harbouring the assessed variant across those disease states represented in the sample dataset. The states selected by the user in the disease states represented in data field will determine whether the familial, sporadic, or affected state rate is requested here. Giving variant characteristics in this format skips Step 1 of the tool workflow (see the 'about' tab) and assumes that the familial/sporadic/affected rate specified is representative of the rate at which that state appears among people who harbour the assessed variant in the population sampled.
If the user has selected to perform error propagation, they should additionally specify either the standard error or the lower confidence interval of this estimate, as previously indicated. If the confidence interval is given, its confidence level must be selected from a list of options; 95% confidence is assumed by default. Note that although the input can accept standard error or confidence intervals, the penetrance estimate output will always be a confidence interval (at the confidence level specified in the confidence level for penetrance estimate field).
Sibship size: The user must specify average sibship size for the sample. This can be given manually or by retrieving estimates of Total Fertility Rate for the sampled population from the World Bank database, provided within the tool under the sibship data repository section, as proxy for sibship size. If drawing data from this repository, the user should select a region and year from the list of options provided. If the query returns a blank response, no fertility rate data were available for the selected region in the selected year; an error message will also be shown. (For additional details, see the 'about' tab).
Disease risk if no variant: The user can opt to specify an indication of lifetime probability of developing disease among family members not harbouring the variant. This defaults to 0, making the assumption that only people harbouring the variant can develop disease. In common traits (e.g where the probability is >0.01) this can bias penetrance estimation. Providing an estimate for this probability is recommended to improve estimation accuracy
Confidence level for penetrance estimate: Where error propagation is performed, the user must specify the confidence level for the output of the tool from a list of options provided in this field field; 95% confidence intervals will be given by default.
Tool output:
Results are presented in a table upon clicking calculate.
Table:
This first presents the 'observed' rate of state X (either familial, sporadic, or affected), calculated from the user data (see Step 1 under the ‘about’ tab). If error propagation is performed, upper and lower confidence intervals and the standard error of this estimate will be provided. If variant characteristics are given in format (3) then the observed rate shown will be the value specified by the user.
The rate of state X within the lookup table that is closest to the observed estimate is taken as the expected rate of X (see Steps 2 and 3). The unadjusted penetrance estimate shown corresponds to this expected rate, at the specified sibship size. The adjusted penetrance estimate should be taken as the final penetrance estimate, and has been obtained by correcting the unadjusted estimate for systematic bias within the approach (see Step 4). If error propagation is performed, confidence intervals are given for the expected disease state rate and these penetrance estimates.
Note that if the observed and expected disease rate values are not approximately equal and penetrance is 0 or 1, is it because the observed disease state rate exceeds or is lesser than the rate expected between an unadjusted penetrance value of 0 (not penetrant) and 1 (fully penetrant) at the specified sibship size. Such an outcome suggests that the variant is either not penetrant for the given outcome or fully penetrant, respective to whether the penetrance estimate is 0 or 1.
An example of usage of the ADPenetrance tool is provided on this tab, illustrating penetrance estimation of SOD1 risk variants for amyotrophic lateral sclerosis. This is estimated based on variant frequencies reported in the familial and sporadic disease states for a European sample of people with ALS collected within a 2017 meta-analysis (Zou et al., 2017).
Each element of tool operation and the returned output has been annotated, marked by box-colour, to briefly describe tool operation as applied to this example. A more comprehensive description of tool operation is given within the tool operation guide under the 'Penetrance Calculator' tab.
Reference list:
Al-Chalabi, A., & Lewis, C. M. (2011). Modelling the Effects of Penetrance and Family Size on Rates of Sporadic and Familial Disease. Human Heredity, 71 (4): 281-288. doi: 10.1159/000330167
Hughes, I. & Hase, T. (2010). Measurements and their uncertainties: a practical guide to modern error analysis, Oxford University Press.
Spargo, T. P., Opie-Martin, S., Bowles, H., Lewis, C. M., Iacoangeli, A., & Al-Chalabi, A. (2022). Calculating variant penetrance from family history of disease and average family size in population-scale data. Genome Medicine 14, 141. doi: 10.1186/s13073-022-01142-7
World Bank, World Development Indicators. Fertility rate, total (births per woman) Retrieved from http://api.worldbank.org/v2/indicator/SP.DYN.TFRT.IN?downloadformat=csv
Zou, Z-Y., Zhou, Z-R., Che, C-H., Liu, C-Y., He, R-L., & Huang, H-P. (2017). Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis, J Neurol Neurosurg Psychiatry, 88 540-549. doi:10.1136/jnnp-2016-315018





